 Resource Balance Analysis
Resource Balance Analysis
Main | What is RBA | Models | Tools | Contact
RBA models
Below you can find RBA models for a few organisms. RBA models are encoded in an XML format, which is not designed to be human-readable. However, network structure, problem parameters, and simulation results can be visualised as tables. To inspect the models and browse their contents, click on the pictures below. You will see an overview page from where you can download model and simulation results as table files. To display the various tables (for example, describing compounds, reactions, proteins etc), you can click on the corresponding region of the overview graphics or use the dropdown menus on top of the page. The organism icon on top the page will bring you back to the model main page, the larger icon on the left brings you back to this page. See "HowTo" (in the dropdown menus) for details about browsing the models.
Click on pictures to see the models
| Bacillus subtilis (wild type) 
|
Escherichia coli (wild type) 
|
Escherichia coli (semi-autotrophic) 
|
Vibrio natriegens
|
Cupriavidus necator (R. eutropha H16) 
|
|
|
|
|
|
| Minimal cell model |
Minimal cell model (Molenaar 2009) |
|||
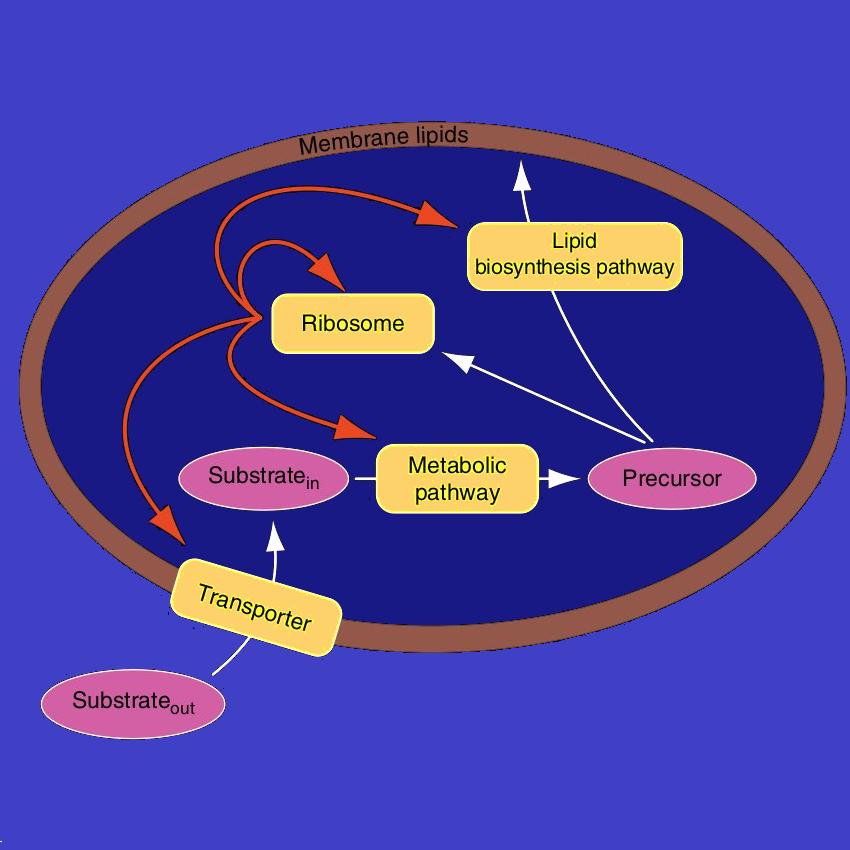
|

|
RBA model download
The following bacterial RBA models (in RBA XML format) can be downloaded from from github:
- Bacillus subtilis 168
- Escherichia coli K-12 (wild type)
- Escherichia coli K-12 (CO2-fixing engineered strain)
- Ralstonia eutropha H16 (also known as Cupriavidus necator)
- Vibrio natriegens (wild-type)
Image copyrights:
Cupriavidus necator: courtesy of Professorship Microbial Biotechnology, TUM