 Resource Balance Analysis
Resource Balance Analysis
Main | What is RBA | Models | Tools | Contact
Overview - Theory - Literature
What is Resource Balance Analysis?
Resource balance analysis (RBA) is an approach for developing models of living cells, assuming that cells use economically their internal resources. The approach considers a biological system at the system level and composed of interacting subsystems; it is therefore a systems biology approach at the intersection of biology, cybernetics and control theory. RBA models accurately capture the macromolecular composition of cells and can be indefinitely refined by adding cellular processes (such as translation, transcription, secretion, chaperoning etc.).
| RBA model structure | RBA model constraints | |
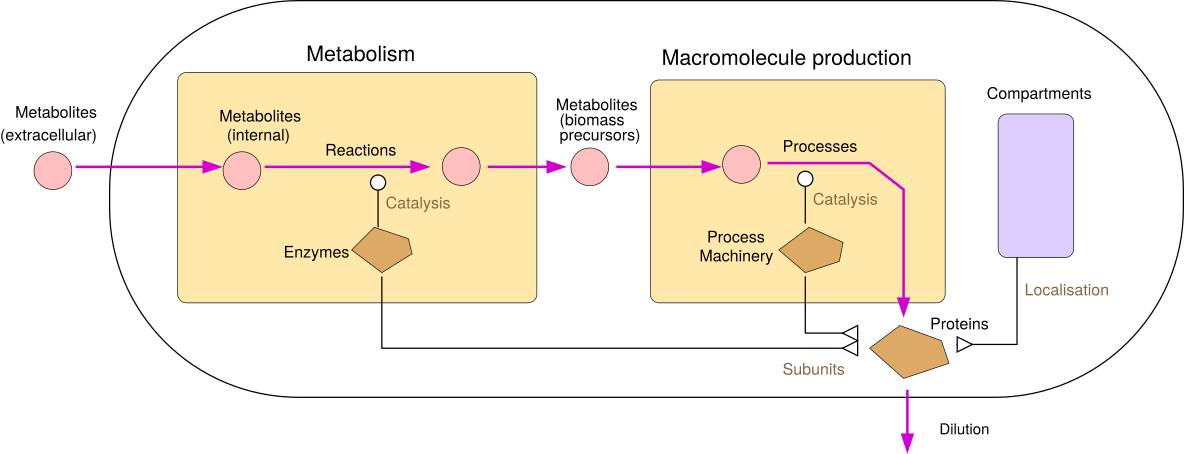
|
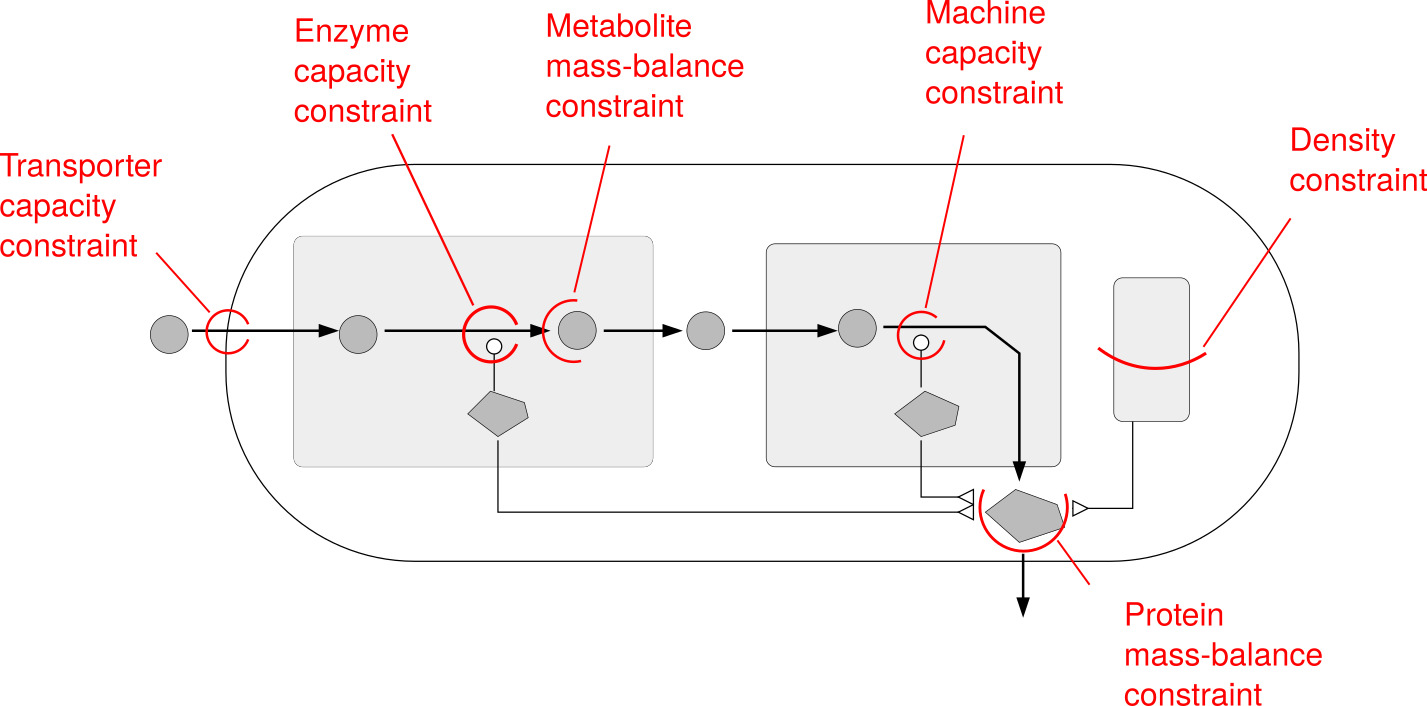
|
The RBA framework, which builds whole-cell models based on metabolic network reconstructions, was developed to describe bacterial populations at steady growth and predefined external nutrient concentrations. It is currently extended to dynamical conditions, to bacterial populations in a chemostat, and to eukaryotic organisms.
But aren't RBA models complicated?
Yes, RBA models are complicated. The reason is that they do not just describe metabolism (like the popular Flux Balance Analysis models), but an entire cell, including many details such as the amino acid composition of proteins, transport processes within a cell, and the composition and efficiency of molecular machines. When building a model, all this knowledge must be gathered. Our tools are made to simplify this process: starting from a pre-existing metabolic model (for instance, a network that would usually be used for Flux Balance Analysis), they add information (about protein sequences and typical molecular machines like the ribosome) automatically, resulting in a first consistent, but fully realistic model. Further details (e.g. other molecular machines, bounds on compartment sizes) can be added manually, but these additional steps require detail knowledge about a cell and cannot be easily simplified. Another step in model building concerns parameter fitting: the efficiencies of enzymes and molecular machines are described by parameters called apparent catalytic constants, which need to be individually determined for each machine, and possibly as a function of cell growth. This estimation can be partially automated, but again involves some manual steps and requires knowledge about the cell to be modelled.
At the end of this model construction, the complete and parameterised model can be stored (and given to colleagues) in a special XML format. All the following steps are less difficult!
How can I run an RBA simulation?
Running simulations with an existing RBA model is fairly easy. Given a model in the RBA XML format, simulations can be run by using the RBApy or RBAtools packages. Pre-built models for different organisms can be downloaded from github. For more details about simulations, see Tools.
How can I build and calibrate an RBA model?
Here we come back to the difficult part. RBA models are built based on existing metabolic network models (the type of models used for Flux Balance Analysis). As explained above, building and calibrating an RBA model by RBApy is a stepwise procedure that is only partially automated, requires manual adjustmenets, and takes some time. For an overview, please see the RBApy package documentation.
Learning more about RBA
- Resource allocation models and RBA are explained in this lecture from the 2022 summer school on Economic Principles in Cell Biology.
- For an introduction to RBApy, see the RBApy homepage and our paper [pdf].
- For an overview of RBAtools, see the online documentation, our paper, and this poster (Metabolic Pathway Analysis 2021).