 Resource Balance Analysis
Resource Balance Analysis
Main | What is RBA | Models | Tools | Contact
Overview - RBAtools - RBApy - RBA online - Downloads
RBAtools - a programming interface for RBA
What is RBAtools?
Building upon RBApy, the RBAtools software package provides an accessible and user-friendly python programming interface to RBA models. It allows users to modify models, contains various high-level simulation and analysis functions, and can export simulation results to different output and visualisation formats. RBApy supports two solvers, the proprietary solver CPLEX and the free solver GLPK. A typical modeling workflow and a general overview of RBAtools are shown below. For a detailed documentation of RBAtools, see the RBAtools user guide and the RBA API reference on github.
RBAtools functionality
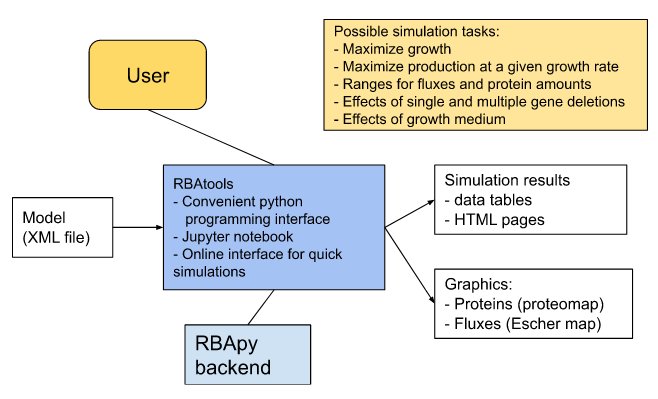
RBAtools provides a python programming interface to RBA models, including some high-level simulation and analysis methods.
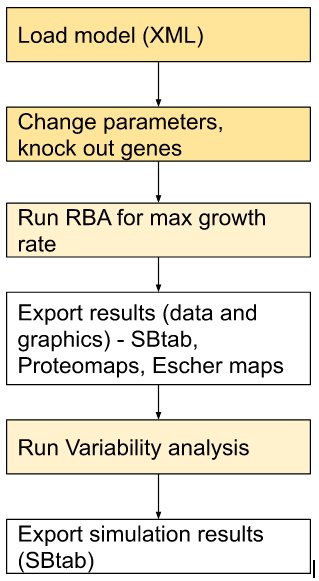
- Easy model manipulation: setting parameters; setting gene knockouts; log, undo, and replay functions
- Data output: output of model and simulation results in a well-structured table format; automatic conversion to HTML
- Data output for visualization tools: metabolic fluxes (Escher maps) and protein amounts (Proteomaps).
- RBAtools can be used directly as a programming interface or through Jupyter notebooks and our online simulator.
| Typical modeling workflow | Information flow in RBAtools | |
 |  |
Model and output formats
Models are imported in the usual RBA XML format (see the RBApy documentation). Models and simulation results can be exported in SBtab formats. RBAtools supports the same output formats for visualization as RBApy: Escher maps (fluxes) and Proteomaps (proteome). In addition, RBA-derived biomass functions for FBA can be exported (see the Jupyter notebooks for details).
RBA simulation in the command line
RBAtools comes with two commands for the Linux command line. To run a RBA simulation, use
run-growth-rate-optimization path/to/rba/model --lp-solver cplex OR swiglpk --output-dir dedicated/location/of/results
To export an RBA model to SBtab format (REF), use
generate-sbtab-of-model-for-html path/to/rba/model --output-dir dedicated/location/of/results
Tutorials
The main functionalities of RBAtools are demonstrated in Jupyter notebooks (details are described here). The RBAtools package on github comes with an example model for B. subtilis bacteria (Goelzer et al. (2015)).
Citing RBAtools
If you use RBAtools for your research, please cite our article Bodeit O. et al. (2023), Bioinformatics Advances, vbad056.
Further information
- Detailed instructions for RBAtools can be found in the RBAtools user guide on github.
- For a detailed description of the RBA code, see the API reference on github.
- For installation and documentation, see Downloads
- An overview of RBAtools is given in this Poster (Metabolic Pathway Analysis 2021)